Automated application of novel gene-disease associations to scale genomic reanalysis
Clinical genomic tests aim to find the cause of a patient’s disease by looking at their DNA. We’ve sequenced the human genome sequence, but we’re still developing our understanding of DNA changes cause disease. Hence, a proprtion of patients won’t receive a diagnosis from genomic testing.
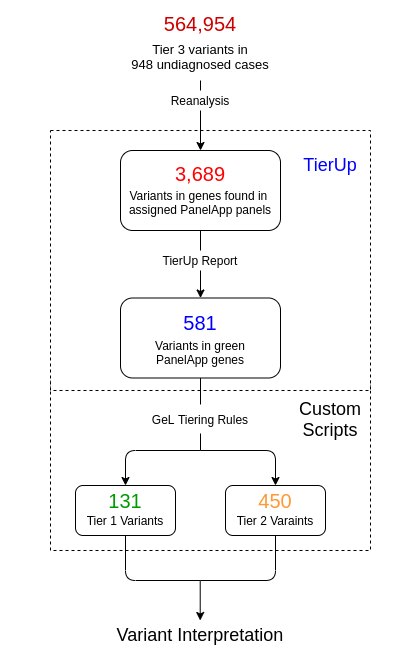
Fortunately genomic data is a digital resource that we can revist, applying new knowlege to diagnose old patients. The difficulty is that doing so is resource intensive, given that new discoveries and the number of unresolved patients both increase with each passing day. To address this issue, we developed TierUp, a bioinformatics tool that searches for new evidence of disease-causing DNA alterations in undiagnosed patients.
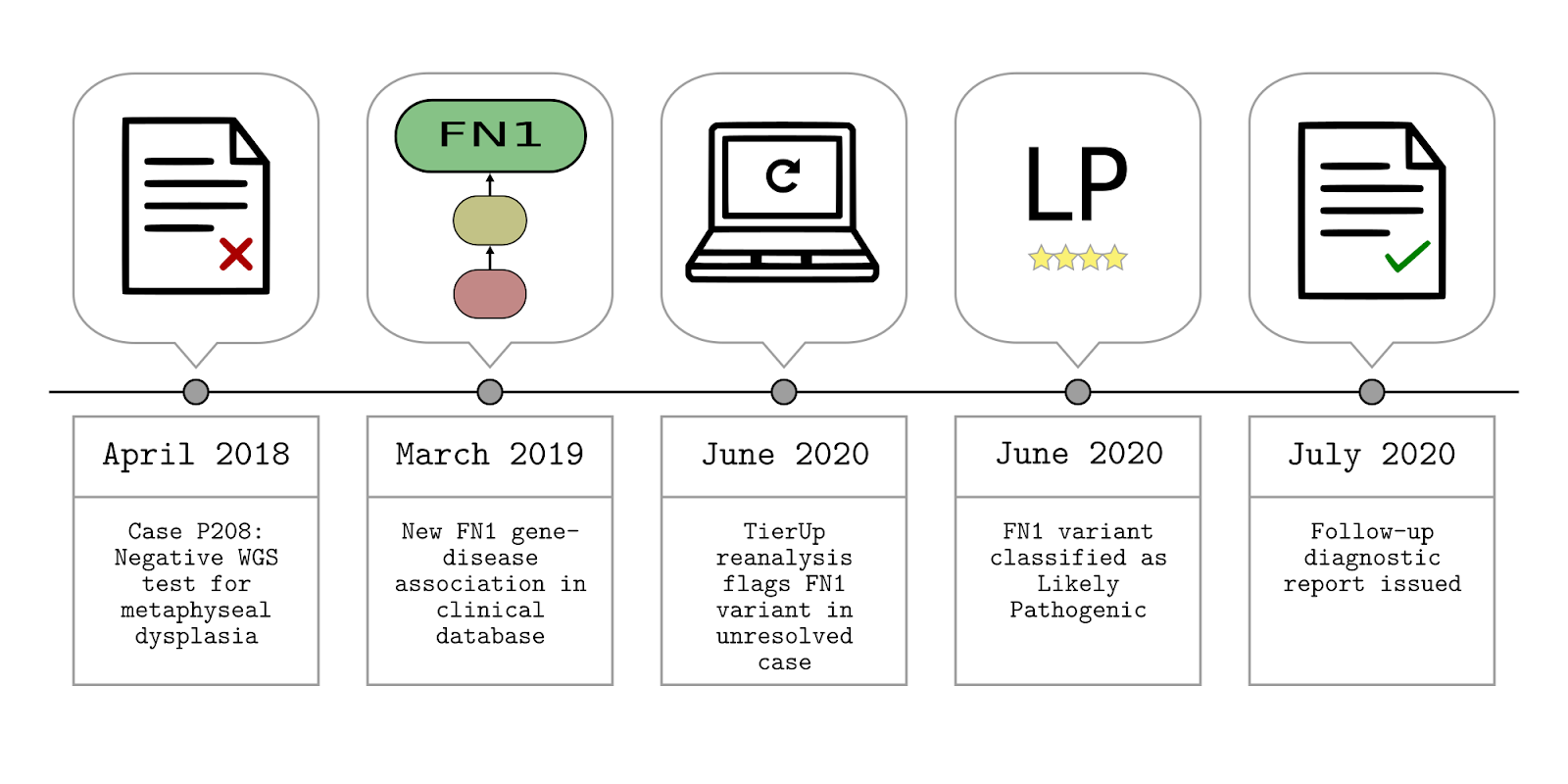
TierUp is specific to patients recruited to the 100,000 genomes project because it provided the infrastructure to automate our method. We used TierUp to reanalyse 948 cases recruited at Guy’s and St Thomas’ NHS Foundation Trust (GSTT), all of whom had yet to receive a diagnosis. We found 410 candidate diagnostic variants in 132 patient cases along with a novel diagnosis.
Our results serve as a proof of principle alongside other studies which demonstrate that bioinformatics tools can streamline reanalysis and lead to additional diagnoses. We encourage NHS labs to use their clinical bioinformatics expertise to develop more open-source tools that will similarly benefit patients.
Our study can be read on medRxiv (not peer-reviewed) and TierUp is available to the community for download.